The goal of this workshop is to not only present the theory and properties of MTAG, but also demonstrate the ease of its application.
Specifically,
- Software built to work from the command line: no programming required
- However, can easily be integrated into Python pipeline (not covered here)
- Organized like the LD Score Regression (
ldsc) tool
- Tutorials and manuals provided on the Github repository
0. Setup¶
(has already been done for you ..)
Install Python¶
- To begin, you need a distribution of Python 2.7 (such as from Anaconda) with the following packages:
numpy,scipy,pandas,argparse,bitarray,joblib.- will also work with Python 3 soon
- You should be able to call Python by typing in
python(or a similar command) in the command line. - Explained in the
READMEfound in the Github repository.
Cloning MTAG from Github¶
- navigate to your folder of choice (
IGSS_2017_mtag_workshopin this demonstration) - use
git clone
- You should now see a folder named
mtagthere
Updating MTAG¶
- navigate into the
mtagfolder - type
git pull
- Up-to-date:
- Otherwise, will update:
1. Basics¶
Summary statistics for this section:¶
| Trait number | Trait | Filename |
|---|---|---|
| 1 | Subjective well-being (SWB) | 1_OA2016_hm3samp_SWB.txt |
| 2 | Neuroticism (NEUR) | 1_OA2016_hm3samp_NEUR.txt |
- Publicly-available sumstats from:
Okbay et al. "Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses." Nature Genetics 48, 624–633 (2016).
- Sample (~10%) of Hapmap 3 SNPs
- Found in the
datafolder
A quick primer on Linux¶
We will only go over the minimum detail needed to follow along in the workshop. There are many good resources for working with Unix available online.
Simple commands¶
cd [path]: change directory to[path](can be relative or absolute)ls [path]: list the files and folder in[path]head -n [N] [file]: view the first[N]lines of[file].taillets you view the last[N]lines.python [path] [-options]: run[path]as a Python script with some options specified. This is how we run MTAG.
Running Bash/Shell scripts¶
- You can write a sequence of commands (and comments) in a
.bashor.shscript bash [file.bash]executes these commands
- This is the easiest way to run the MTAG commands in this tutorial
Listing options (-h)¶
- General format:
python [PATH_OF_MTAG_CODE] -h - For, example in the
mtagfolder:
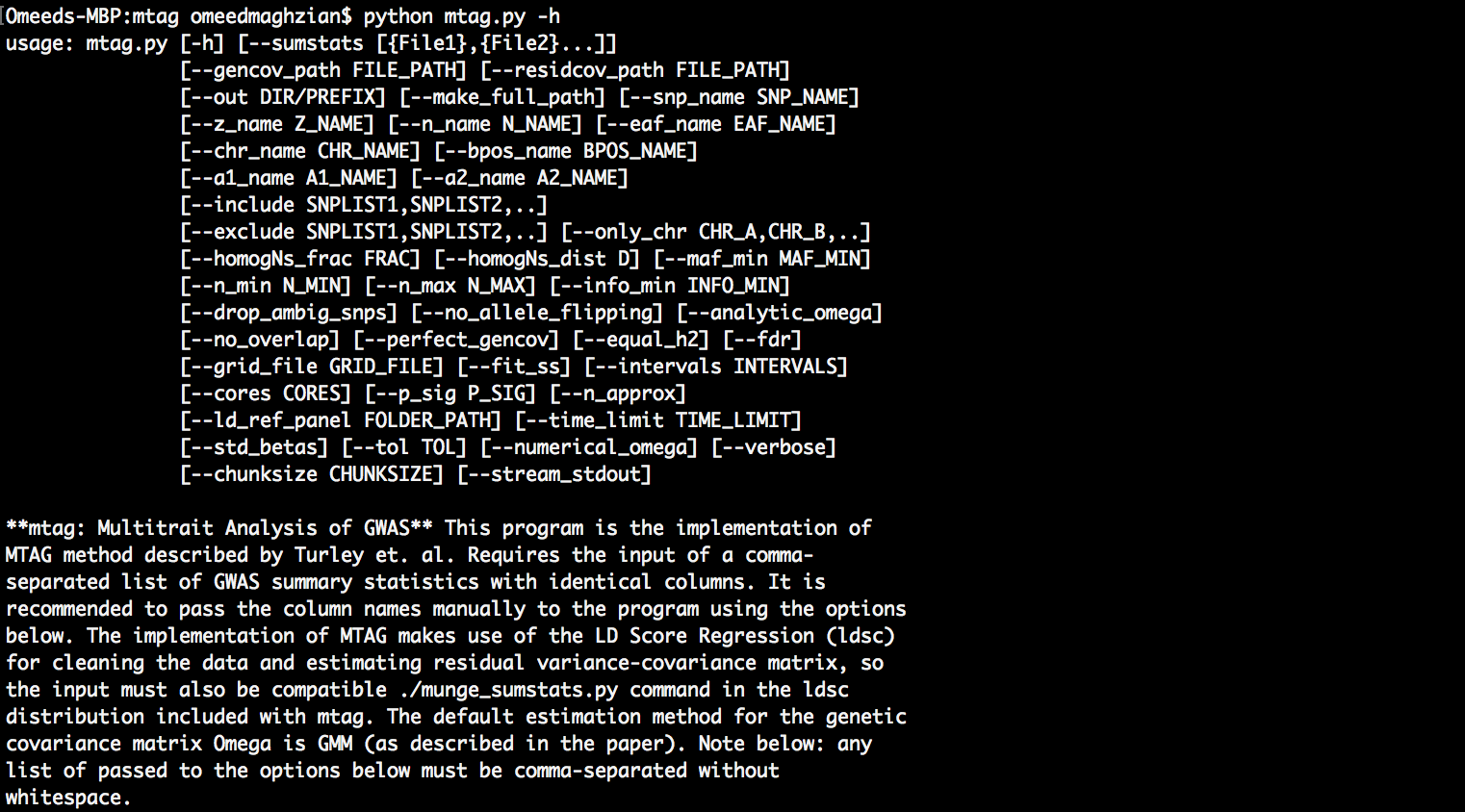
# 1.1_access_help.bash
!python ../mtag/mtag.py -h
usage: mtag.py [-h] [--sumstats [{File1},{File2}...]]
[--gencov_path FILE_PATH] [--residcov_path FILE_PATH]
[--out DIR/PREFIX] [--make_full_path] [--snp_name SNP_NAME]
[--z_name Z_NAME] [--n_name N_NAME] [--eaf_name EAF_NAME]
[--chr_name CHR_NAME] [--bpos_name BPOS_NAME]
[--a1_name A1_NAME] [--a2_name A2_NAME]
[--include SNPLIST1,SNPLIST2,..]
[--exclude SNPLIST1,SNPLIST2,..] [--only_chr CHR_A,CHR_B,..]
[--homogNs_frac FRAC] [--homogNs_dist D] [--maf_min MAF_MIN]
[--n_min N_MIN] [--n_max N_MAX] [--info_min INFO_MIN]
[--drop_ambig_snps] [--no_allele_flipping] [--analytic_omega]
[--no_overlap] [--perfect_gencov] [--equal_h2] [--fdr]
[--grid_file GRID_FILE] [--fit_ss] [--intervals INTERVALS]
[--cores CORES] [--p_sig P_SIG] [--n_approx]
[--ld_ref_panel FOLDER_PATH] [--time_limit TIME_LIMIT]
[--std_betas] [--tol TOL] [--numerical_omega] [--verbose]
[--chunksize CHUNKSIZE] [--stream_stdout]
**mtag: Multitrait Analysis of GWAS** This program is the implementation of
MTAG method described by Turley et. al. Requires the input of a comma-
separated list of GWAS summary statistics with identical columns. It is
recommended to pass the column names manually to the program using the options
below. The implementation of MTAG makes use of the LD Score Regression (ldsc)
for cleaning the data and estimating residual variance-covariance matrix, so
the input must also be compatible ./munge_sumstats.py command in the ldsc
distribution included with mtag. The default estimation method for the genetic
covariance matrix Omega is GMM (as described in the paper). Note below: any
list of passed to the options below must be comma-separated without
whitespace.
optional arguments:
-h, --help show this help message and exit
Input Files:
Input files to be used by MTAG. The --sumstats option is required, while
using the other two options take priority of their corresponding
estimation routines, if used.
--sumstats [{File1},{File2}...]
Specify the list of summary statistics files to
perform multitrait analysis. Multiple files paths must
be separated by ",". Please read the documentation to
find the up-to-date set of acceptable file formats. A
general guideline is that any files you pass into MTAG
should also be parsable by ldsc and you should take
the additional step of specifying the names of the
main columns below to avoid reading errors.
--gencov_path FILE_PATH
If specified, will read in the genetic covariance
matrix saved in the file path below and skip the
estimation routine. The rows and columns of the matrix
must correspond to the order of the GWAS input files
specified. FIles can either be in whitespace-delimited
.txt or .npy format. Use with caution as the genetic
covariance matrix specified will be weakly nonoptimal.
--residcov_path FILE_PATH
If specified, will read in the residual covariance
matrix saved in the file path below and skip the
estimation routine. The rows and columns of the matrix
must correspond to the order of the GWAS input files
specified. FIles can either be in .txt or .npy format.
Use with caution as the genetic covariance matrix
specified will be weakly nonoptimal. File must either
be in whitespace-delimited .txt or .npy
Output formatting:
Set the output directory and common name of prefix files.
--out DIR/PREFIX Specify the directory and name prefix to output MTAG
results. All mtag results will be prefixed with the
corresponding tag. Default is ./mtag_results
--make_full_path option to make output path specified in -out if it
does not exist.
Column names of input files:
These options manually pass the names of the relevant summary statistics
columns used by MTAG. It is recommended to pass these names because only
narrow searches for these columns are performed in the default cases.
Moreover, it is necessary that these input files be readable by ldsc's
munge_sumstats command.
--snp_name SNP_NAME Name of the single column that provides the unique
identifier for SNPs in the GWAS summary statistics
across all GWAS results. Default is "snpid". This the
index that will be used to merge the GWAS summary
statistics. Any SNP lists passed to ---include or
--exclude should also contain the same name.
--z_name Z_NAME The common name of the column of Z scores across all
input files. Default is to search for columns
beginning with the lowercase letter z.
--n_name N_NAME the common name of the column of sample sizes in the
GWAS summary statistics files. Default is to search
for columns beginning with the lowercase letter n.
--eaf_name EAF_NAME The common name of the column of minor allele
frequencies (MAF) in the GWAS input files. The default
is "freq".
--chr_name CHR_NAME Name of the column containing the chromosome of each
SNP in the GWAS input. Default is "chr".
--bpos_name BPOS_NAME
Name of the column containing the base pair of each
SNP in the GWAS input. Default is "bpos".
--a1_name A1_NAME Name of the column containing the effect allele of
each SNP in the GWAS input. Default is "a1".
--a2_name A2_NAME Name of the column containing the non-effect allele of
each SNP in the GWAS input. Default is "a2".
Filter Options:
The input summary statistics files can be filtered using the options
below. Note that there is some default filtering according to sample size
and allele frequency, following the recommendations we make in the
corresponding paper. All of these column-based options allow a list of
values to be passed of the same length as the number of traits
--include SNPLIST1,SNPLIST2,..
Restricts MTAG analysis to the union of snps in the
list of snplists provided. The header line must match
the SNP index that will be used to merge the GWAS
input files.
--exclude SNPLIST1,SNPLIST2,.., --excludeSNPs SNPLIST1,SNPLIST2,..
Similar to the --include option, except that the union
of SNPs found in the specified files will be excluded
from MTAG. Both -exclude and -include may be
simultaneously specified, but -exclude will take
precedent (i.e., SNPs found in both the -include and
-exclude SNP lists will be excluded).
--only_chr CHR_A,CHR_B,..
Restrict MTAG to SNPs on one of the listed, comma-
separated chromosome. Can be specified simultaneously
with --include and --exclude, but will take precedent
over both. Not generally recommended. Multiple
chromosome numbers should be separated by commas
without whitespace. If this option is specified, the
GWAS summary statistics must also list the chromosome
of each SNPs in a column named \`chr\`.
--homogNs_frac FRAC Restricts to SNPs within FRAC of the mode of sample
sizes for the SNPs as given by (N-Mode)/Mode < FRAC.
This filter is not applied by default.
--homogNs_dist D Restricts to SNPs within DIST (in sample size) of the
mode of sample sizes for the SNPs. This filter is not
applied by default.
--maf_min MAF_MIN set the threshold below SNPs with low minor allele
frequencies will be dropped. Default is 0.01. Set to 0
to skip MAF filtering.
--n_min N_MIN set the minimum threshold for SNP sample size in input
data. Default is 2/3*(90th percentile). Any SNP that
does not pass this threshold for all of the GWAS input
statistics will not be included in MTAG.
--n_max N_MAX set the maximum threshold for SNP sample size in input
data. Not used by default. Any SNP that does not pass
this threshold for any of the GWAS input statistics
will not be included in MTAG.
--info_min INFO_MIN Minimim info score for filtering SNPs for MTAG.
--drop_ambig_snps Drop strand ambiguous SNPs when performing MTAG (they
are already not used to estimate Omega or Sigma.
--no_allele_flipping Prevents flipping the effect sizes of summary
statistics when the effect and non-effect alleles are
reversed (reletive the first summary statistics file.
Special Cases:
These options deal with notable special cases of MTAG that yield
improvements in runtime. However, they should be used with caution as they
will yield non-optimal results if the assumptions implicit in each option
are violated.
--analytic_omega Option to turn off the numerical estimation of the
genetic VCV matrix in the presence of constant sample
size within each GWAS, for which a closed-form
solution exists. The default is to typically use the
closed form solution as the starting point for the
numerical solution to the maximum-likelihood genetic
VCV, Use with caution! If any input GWAS does not have
constant sample size, then the analytic solution
employed here will not be a maximizer of the
likelihood function.
--no_overlap Imposes the assumption that there is no sample overlap
between the input GWAS summary statistics. MTAG is
performed with the off-diagonal terms on the residual
covariance matrix set to 0.
--perfect_gencov Imposes the assumption that all traits used are
perfectly genetically correlated with each other. The
off-diagonal terms of the genetic covariance matrix
are set to the square root of the product of the
heritabilities
--equal_h2 Imposes the assumption that all traits passed to MTAG
have equal heritability. The diagonal terms of the
genetic covariance matrix are set equal to each other.
Can only be used in conjunction with --perfect_gencov
Max FDR calculation:
These options are used for the calculation of an upper bound on the false
disovery under the model described in Supplementary Note 1.1.4 of Turley
et al. (2017). Note that there is one of three ways to define the space of
grid points over which the upper bound is searched.
--fdr Perform max FDR calculations
--grid_file GRID_FILE
Pre-set list of grid points. Users can define a list
of grid points over which the search is conducted. The
list of grid points should be passed in text file as a
white-space delimited matrix of dimnesions, G x S,
where G is the number of grid points and S = 2^T is
the number of possible causal states for SNPs. States
are ordered according to a tree-like recursive
structure from right to left. For example, for 3
traits, with the triple TFT denoting the state for
which SNPs are causal for State 1, not causal for
state 2, and causal for state 3, then the column
ordering of probabilities should be: FFF FFT FTF FTT
TFF TFT TTF TTT There should be no headers, or row
names in the file. Any rows for which (i) the
probabilities do not sum to 1, the prior of a SNP
being is causal is 0 for any of the traits, and (iii)
the resulting genetic correlation matrix is non
positive definite will excluded in the search.
--fit_ss This estimates the prior probability that a SNP is
null for each trait and then proceeds to restrict the
grid search to the set of probability vectors that sum
to the prior null for each trait. This is useful for
restrict the search space of larger-dimensional
traits.
--intervals INTERVALS
Number of intervals that you would like to partition
the [0,1] interval. For example example, with two
traits and --intervals set 10, then maxFDR will
calculated over the set of feasible points in {0.,
0.1, 0.2,..,0.9,1.0}^2.
--cores CORES Number of threads/cores use to compute the FDR grid
points for each trait.
--p_sig P_SIG P-value threshold used for statistical signifiance.
Default is p=5.0e-8 (genome-wide significance).
--n_approx Speed up FDR calculation by replacing the sample size
of a SNP for each trait by the mean across SNPs (for
each trait). Recommended.
Winner's curse adjustment:
Options related to the winner's curse adjustment of estimates of effect
sizes from MTAG that could be used when replicating analyses.
Miscellaneous:
--ld_ref_panel FOLDER_PATH
Specify folder of the ld reference panel (split by
chromosome) that will be used in the estimation of the
error VCV (sigma). This option is passed to --ref-ld-
chr and --w-ld-chr when running LD score regression.
The default is to use the reference panel of LD scores
computed from 1000 Genomes European subjects
(eur_w_ld_chr) that is included with the distribution
of MTAG
--time_limit TIME_LIMIT
Set time limit (hours) on the numerical estimation of
the variance covariance matrix for MTAG, after which
the optimization routine will complete its current
iteration and perform MTAG using the last iteration of
the genetic VCV.
--std_betas Results files will have standardized effect sizes,
i.e., the weights 1/sqrt(2*MAF*(1-MAF)) are not
applied when outputting MTAG results, where MAF is the
minor allele frequency.
--tol TOL Set the relative (x) tolerance when numerically
estimating the genetic variance-covariance matrix. Not
recommended to change unless you are facing strong
runtime constraints for a large number of traits.
--numerical_omega Option to use the MLE estimator of the genetic VCV
matrix, implemented through a numerical routine.
--verbose When used, will include output from running ldsc
scripts as well additional information (such as
optimization routine information.
--chunksize CHUNKSIZE
Chunksize for reading in data.
--stream_stdout Will streat mtag processing on console in addition to
writing to log file.
A. The two options that must always be specified¶
--sumstats [File1],[File2],..- Comma-separated list of summary statistics files to apply MTAG. Currently, files must be whitespace-delimited and must either have the default column names or column names that are provided through options (see below).
- Should also make sure that files can be read by
ldsc(included in themtagfolder)
--out [output_folder][prefix]- The path provided consists of a folder
[output_folder](e.g.,../output/or./) - The
[prefix](e.g.,igss_mtag_tutorial_) is prepended to the files produced by MTAG
- The path provided consists of a folder
B. Sumstats file formatting¶
- The column names must be consistent across all sumstats files.
- At present
mtagdoes not "guess" the column names (will hopefully do that soon). - Column order does not matter.
- All files must be whitespace (tab) delimited
- At present
The workshop files provided contain the "default" column names for MTAG
bash 1.2_view_sumstats.bash¶
# 1.2 view sample sumstats files (for format)
!head ../data/1_OA2016_hm3samp_NEUR.txt # 1.2
snpid chr bpos a1 a2 freq z pval n rs2736372 8 11106041 T C 0.4179 -7.71614161262 1.209e-14 111111.111111 rs2060465 8 11162609 T C 0.6194 7.69444599845 1.422e-14 62500.0 rs10096421 8 10831868 T G 0.4646 -7.561098219 3.989e-14 111111.111111 rs2409722 8 11039816 T G 0.4627 -7.38261601808 1.553e-13 111111.111111 rs11991118 8 10939273 T G 0.5056 7.32202915636 2.443e-13 111111.111111 rs2736371 8 11105529 A G 0.3806 -7.32009158327 2.478e-13 62500.0 rs2736313 8 11086942 T C 0.4646 -7.24228035161 4.411e-13 111111.111111 rs876954 8 8310923 A G 0.4813 -7.15791677597 8.191e-13 62500.0 rs1533059 8 8684953 A G 0.4478 -7.07412856424 1.504e-12 62500.0
All of the columns shown above are necessary for MTAG to run
!tail -5 ../data/1_OA2016_hm3samp_NEUR.txt
rs165119 18 75317995 T C 0.1101 0.0 1.0 27777.7777778 rs1916337 3 165252023 T C 0.2444 0.0 1.0 62500.0 rs4878187 9 38227021 T C 0.8713 0.0 1.0 40000.0 rs8080313 17 35952983 A G 0.8396 0.0 1.0 40000.0 rs851997 6 151985550 A G 0.3787 0.0 1.0 62500.0
!sed -n '103995,104000p' ../data/1_OA2016_hm3samp_NEUR.txt
rs322382 5 172198952 T C 0.3041 -0.648595291061 0.5166 62500.0 rs3795732 1 156564640 A G 0.7108 -0.648595291061 0.5166 62500.0 rs4111166 9 26524416 T C 0.4739 0.648595291061 0.5166 111111.111111 rs4491677 2 128789204 A G 0.2276 -0.648595291061 0.5166 62500.0 rs622272 17 14400878 T G 0.5 -0.648595291061 0.5166 111111.111111 rs842938 2 201404694 T C 0.459 0.648595291061 0.5166 111111.111111
Running MTAG with the defaults¶
Using mtag with the default options implements the following steps:
- Read in each summary statistics file in the list and "munges" (filters/clean) them, in particular MTAG:
- Only keeps SNPs with Minor Allele Frequency (MAF) $ \geq 0.01$
- Only keeps SNPs with sample size $N \geq \frac{2}{3} F^{-1}_N(90)$ (greater than two-thirds of the 90th percentile)
- Merge the filtered GWAS summary statistics results together, taking the intersection of available SNPs (accounting for lists of SNPs to
includeand/orexclude). - Estimate the residual covariance matrix, $\Sigma_{LD}$, using the intercept of LD Score regression (
ldsc, longest step). - Estimate the genetic covariance matrix, $\Omega$.
- Calculate MTAG-adjusted statistics and output results.
Running MTAG with the defaults¶
Call the mtag.py script via python
Note: we use the --stream_stdout so that messages from MTAG are printed to the console in addition to the log file.
bash 1.3_mtag_default.bash¶
# 1.3_mtag_default.bash
!python ../mtag/mtag.py \
--sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \
--out ../output/1_basics/1.3_mtag_default_NS \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.3 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --stream-stdout \ --sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \ --out ../output/1_basics/1.3_mtag_default_NS Beginning MTAG analysis... Read in Trait 1 summary statistics (185458 SNPs) from ../data/1_OA2016_hm3samp_NEUR.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 185458 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 185458 SNPs remain. Removed 0 SNPs with duplicated rs numbers (185458 SNPs remain). Removed 162680 SNPs with N < 74074.074074 (22778 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.467 Lambda GC = 1.366 Max chi^2 = 59.523 20 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Thu Oct 19 02:03:44 2017 Total time elapsed: 0.7s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 22778 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (167911 SNPs) from ../data/1_OA2016_hm3samp_SWB.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 167911 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 167911 SNPs remain. Removed 0 SNPs with duplicated rs numbers (167911 SNPs remain). Removed 102537 SNPs with N < 74074.074074 (65374 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.208 Lambda GC = 1.188 Max chi^2 = 29.353 1 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Thu Oct 19 02:03:46 2017 Total time elapsed: 1.21s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 65374 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 20321 SNPs remaining merging with previous traits. ... Merge of GWAS summary statistics complete. Number of SNPs: 20321 Using 20321 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 1.043 -0.129] [-0.129 0.966]] Beginning estimation of Omega ... Using GMM estimator of Omega .. Checking for positive definiteness .. Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 ...hm3samp_NEUR.txt 111111 111111 20321 1.407 1.488 133309 2 ..._hm3samp_SWB.txt 111111 111111 20321 1.269 1.408 168490 Estimated Omega: [[ 3.817e-06 -2.199e-06] [ -2.199e-06 2.336e-06]] Estimated Sigma: [[ 1.043 -0.129] [-0.129 0.966]] MTAG results saved to file. MTAG complete. Time elapsed: 27.0710499287s
Interpreting Output¶
Reading the log file: masthead¶
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
<>
<> MTAG: Multitrait Analysis of GWAS
<> Version: 1.0.1
<> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley
<> Harvard University Department of Economics / Broad Institute of MIT and Harvard
<> GNU General Public License v3
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
<> Note: It is recommended to run your own QC on the input before using this program.
<> Software-related correspondence: maghzian@nber.org
<> All other correspondence: paturley@broadinstitute.org<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
Calling ./mtag.py \
--stream-stdout \
--sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \
--out ../output/part_2/igss_mtag_default_NS
Beginning MTAG analysis...Reading the log file: munging¶
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
Interpreting column names as follows:
snpid: Variant ID (e.g., rs number)
n: Sample size
a1: Allele 1, interpreted as ref allele for signed sumstat.
pval: p-Value
a2: Allele 2, interpreted as non-ref allele for signed sumstat.
freq: Allele frequency
z: Directional summary statistic as specified by --signed-sumstats.
...
...
...Reading the log file: estimation¶
...
... Merge of GWAS summary statistics complete. Number of SNPs: 20321
Using 20321 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity)
Estimating sigma..
Checking for positive definiteness ..
Sigma hat:
[[ 1.043 -0.129]
[-0.129 0.966]]
Beginning estimation of Omega ...
Using GMM estimator of Omega ..
Checking for positive definiteness ..
Completed estimation of Omega ...
Beginning MTAG calculations...
... Completed MTAG calculations.
Writing Phenotype 1 to file ...
Writing Phenotype 2 to file ...
...Reading the Log File (Summary Output)¶
Summary of MTAG results:
------------------------
Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N
1 ...hm3samp_NEUR.txt 111111 111111 20321 1.407 1.488 133309
2 ..._hm3samp_SWB.txt 111111 111111 20321 1.269 1.408 168490
Estimated Omega:
[[ 3.817e-06 -2.199e-06]
[ -2.199e-06 2.336e-06]]
Estimated Sigma:
[[ 1.043 -0.129]
[-0.129 0.966]](genetic correlation estimated by MTAG)¶
[[ 1. , -0.7363645],
[-0.7363645, 1. ]]Results files¶
bash 1.3b_list_files.bash¶
# 1.3b_list_files.bash
!ls ../output/1_basics/1.3*
../output/1_basics/1.3_mtag_default_NS.log ../output/1_basics/1.3_mtag_default_NS_omega_hat.txt ../output/1_basics/1.3_mtag_default_NS_sigma_hat.txt ../output/1_basics/1.3_mtag_default_NS_trait_1.txt ../output/1_basics/1.3_mtag_default_NS_trait_2.txt
Results files¶
T + 3 files (T number of input sumstats), all whitespace-delimited
*.log: Log file (shown above)*_omega_hat.txt: Estimated genetic covariance matrix*_sigma_hat.txt: Estimated residual covariance matrix
*_trait_[t].txt: MTAG results for trait $t=1,\ldots, T$- Let's look at these files a bit more ...
bash 1.3c_view_mtag_output.bash¶
!head "../output/1_basics/1.3_mtag_default_NS_trait_1.txt"
snpid chr bpos a1 a2 z n freq mtag_beta mtag_se mtag_z mtag_pval rs2736372 8 11106041 T C -7.71614161262 111111.111111 0.4179 -0.0312843727759 0.00401008053375 -7.80143254297 6.12083232605e-15 rs10096421 8 10831868 T G -7.561098219 111111.111111 0.4646 -0.0292358562051 0.0039656035404 -7.37235982046 1.67633824925e-13 rs2409722 8 11039816 T G -7.38261601808 111111.111111 0.4627 -0.0314168522186 0.00396670505601 -7.92013819404 2.37246749e-15 rs11991118 8 10939273 T G 7.32202915636 111111.111111 0.5056 0.0295263257704 0.00395590010421 7.46387041953 8.40172307848e-14 rs2736313 8 11086942 T C -7.24228035161 111111.111111 0.4646 -0.0296988957677 0.0039656035404 -7.48912377779 6.93349296434e-14 rs7016385 8 10779472 T C 7.014366598 111111.111111 0.5373 0.0259293691662 0.00396670505601 6.53675249359 6.28689598667e-11 rs11250130 8 11214455 A G 6.609661126 111111.111111 0.4869 0.026363608706 0.00395701034061 6.66250690212 2.69195672093e-11 rs7001819 8 11650475 T C -6.40845674838 111111.111111 0.5728 -0.0237731953794 0.00399825927266 -5.94588638658 2.74964820715e-09 rs4240671 8 10767748 A G 6.35561088027 111111.111111 0.4981 0.0233347188014 0.00395568054249 5.89904026646 3.65622009607e-09
| snpid | a1 | a2 | z | mtag_beta | mtag_se | mtag_z | mtag_pval |
|---|---|---|---|---|---|---|---|
| rs2736372 | T | C | -7.71614161262 | -0.0312843727759 | 0.00401008053375 | -7.80143254297 | 6.12083232605e-15 |
| rs10096421 | T | G | -7.561098219 | -0.0292358562051 | 0.0039656035404 | -7.37235982046 | 1.67633824925e-13 |
| rs2409722 | T | G | -7.38261601808 | -0.0314168522186 | 0.00396670505601 | -7.92013819404 | 2.37246749e-15 |
| rs11991118 | T | G | 7.32202915636 | 0.0295263257704 | 0.00395590010421 | 7.46387041953 | 8.40172307848e-14 |
| rs2736313 | T | C | -7.24228035161 | -0.0296988957677 | 0.0039656035404 | -7.48912377779 | 6.93349296434e-14 |
| snpid | a1 | a2 | z | mtag_beta | mtag_se | mtag_z | mtag_pval |
|---|---|---|---|---|---|---|---|
| rs2736372 | T | C | -7.71614161262 | -0.0312843727759 | 0.00401008053375 | -7.80143254297 | 6.12083232605e-15 |
| rs10096421 | T | G | -7.561098219 | -0.0292358562051 | 0.0039656035404 | -7.37235982046 | 1.67633824925e-13 |
| rs2409722 | T | G | -7.38261601808 | -0.0314168522186 | 0.00396670505601 | -7.92013819404 | 2.37246749e-15 |
| rs11991118 | T | G | 7.32202915636 | 0.0295263257704 | 0.00395590010421 | 7.46387041953 | 8.40172307848e-14 |
| rs2736313 | T | C | -7.24228035161 | -0.0296988957677 | 0.0039656035404 | -7.48912377779 | 6.93349296434e-14 |
Some comments:¶
- Only contains SNPs the survive all filters
- Ordered the same across traits
- All columns beginning with
mtag_is produced by MTAG - Default
mtag_betaandmtag_seare unstandardized by genotype- Use the
--std_betasoptions to produce standardized beta/se
- Use the
- Allele columns are aligned across traits (matched to orientation of first input file)
Warning: low sample might lead to non-positive definite estimates of $\Omega$, $\Sigma_{LD}$ (truncated by program)!¶
Column name options¶
| Column | option | Default |
|---|---|---|
| SNP ID | --snp_name |
snpid |
| Chromosome | --chr_name |
chr |
| Base-pair position | --bpos_name |
bpos |
| Z-score | --z_name |
z* |
| sample size | --n_name |
n |
| expected allele frequency | --eaf_name |
freq |
| Allele 1 (effect allele) | --a1_name |
a1 |
| Allele 2 (other allele) | --a2_name |
a2 |
Specifying matrix paths¶
- Use:
--gencov_pathto specify $\Omega$--residcov_pathto specify $\Sigma_{LD}$
- Files must be in the same format as produced by MTAG (contain only the matrix).
Let's try it out . . .¶
bash 1.4_mtag_matrices.bash¶
!python ../mtag/mtag.py \
--sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \
--out ../output/1_basics/1.4_mtag_withMat_NS \
--gencov_path ../output/1_basics/1.3_mtag_default_NS_omega_hat.txt \
--residcov_path ../output/1_basics/1.3_mtag_default_NS_sigma_hat.txt \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.2 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --gencov-path ../output/1_basics/1.3_mtag_default_NS_omega_hat.txt \ --stream-stdout \ --residcov-path ../output/1_basics/1.3_mtag_default_NS_sigma_hat.txt \ --sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \ --out ../output/1_basics/1.4_mtag_withMat_NS Beginning MTAG analysis... Read in Trait 1 summary statistics (185458 SNPs) from ../data/1_OA2016_hm3samp_NEUR.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 185458 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 185458 SNPs remain. Removed 0 SNPs with duplicated rs numbers (185458 SNPs remain). Removed 162680 SNPs with N < 74074.074074 (22778 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.467 Lambda GC = 1.366 Max chi^2 = 59.523 20 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 16:09:14 2017 Total time elapsed: 0.61s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 22778 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (167911 SNPs) from ../data/1_OA2016_hm3samp_SWB.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 167911 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 167911 SNPs remain. Removed 0 SNPs with duplicated rs numbers (167911 SNPs remain). Removed 102537 SNPs with N < 74074.074074 (65374 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.208 Lambda GC = 1.188 Max chi^2 = 29.353 1 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 16:09:15 2017 Total time elapsed: 0.74s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 65374 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 20321 SNPs remaining merging with previous traits. ... Merge of GWAS summary statistics complete. Number of SNPs: 20321 Using 20321 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Checking for positive definiteness .. Sigma hat: [[ 1.043 -0.129] [-0.129 0.966]] Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 ...hm3samp_NEUR.txt 111111 111111 20321 1.407 1.488 133309 2 ..._hm3samp_SWB.txt 111111 111111 20321 1.269 1.408 168490 Estimated Omega: [[ 3.817e-06 -2.199e-06] [ -2.199e-06 2.336e-06]] Estimated Sigma: [[ 1.043 -0.129] [-0.129 0.966]] MTAG results saved to file. MTAG complete. Time elapsed: 3.93074512482s
Filtering: maintain $\Omega$, $\Sigma_j$ assumptions¶
2. Special Options¶
- See Online methods of paper for detailed description
- Be cognizant of the assumptions implicit in each option!
Sumstats we will be using¶
- EA3 Meta-analysis (Okbay et al. 2016):
EducAtt_ea3.txt - Edu_years GWAS with UKB interim release:
EducAtt_ukb.txt
Without special options:¶
!python ../mtag/mtag.py \
--sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \
--out ../output/2_special_opts/2.0_mtag_EA \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.2 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --stream-stdout \ --sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \ --out ../output/2_special_opts/2.0_mtag_EA Beginning MTAG analysis... Read in Trait 1 summary statistics (8146840 SNPs) from ../data/EducAtt_ea2.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 8146840 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 8146840 SNPs remain. Removed 0 SNPs with duplicated rs numbers (8146840 SNPs remain). Removed 4434138 SNPs with N < 74074.074074 (3712702 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.85 Lambda GC = 1.627 Max chi^2 = 106.943 5297 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:12:58 2017 Total time elapsed: 34.62s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 3712702 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (1191004 SNPs) from ../data/EducAtt_ukb.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 1191004 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 12960 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 1178044 SNPs remain. Removed 0 SNPs with duplicated rs numbers (1178044 SNPs remain). Removed 0 SNPs with N < 35242.0 (1178044 SNPs remain). Median value of SIGNED_SUMSTATS was -0.000292861940766, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.205 Lambda GC = 1.192 Max chi^2 = 39.518 23 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:13:23 2017 Total time elapsed: 5.41s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 1178044 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 790847 SNPs remaining merging with previous traits. Dropped 1 SNPs due to inconsistent allele pairs from phenotype 2. 790846 SNPs remain. Flipped the signs of of 384828 SNPs to make them consistent with the effect allele orderings of the first trait. ... Merge of GWAS summary statistics complete. Number of SNPs: 790846 Using 790846 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 0.92 0.385] [ 0.385 1.028]] Beginning estimation of Omega ... Using GMM estimator of Omega .. Checking for positive definiteness .. Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 2.022 252546 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 1.841 217709 Estimated Omega: [[ 6.492e-06 4.815e-06] [ 4.815e-06 3.970e-06]] Estimated Sigma: [[ 0.92 0.385] [ 0.385 1.028]] MTAG results saved to file. MTAG complete. Time elapsed: 2.0m:58.9194750786s
!tail -26 ../output/2_special_opts/2.0_mtag_EA.log
2017/10/18/06:14:31 PM Beginning estimation of Omega ... 2017/10/18/06:14:31 PM Using GMM estimator of Omega .. 2017/10/18/06:14:31 PM Checking for positive definiteness .. 2017/10/18/06:14:31 PM Completed estimation of Omega ... 2017/10/18/06:14:31 PM Beginning MTAG calculations... 2017/10/18/06:14:32 PM ... Completed MTAG calculations. 2017/10/18/06:14:32 PM Writing Phenotype 1 to file ... 2017/10/18/06:14:49 PM Writing Phenotype 2 to file ... 2017/10/18/06:15:07 PM Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 2.022 252546 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 1.841 217709 Estimated Omega: [[ 6.492e-06 4.815e-06] [ 4.815e-06 3.970e-06]] Estimated Sigma: [[ 0.92 0.385] [ 0.385 1.028]] 2017/10/18/06:15:07 PM 2017/10/18/06:15:07 PM MTAG results saved to file. 2017/10/18/06:15:07 PM MTAG complete. Time elapsed: 2.0m:58.9194750786s
--no_overlap: MTAG in the absence of sample overlap¶
Assumes: no overlap between any of the cohorts in any pair of GWAS sumstats¶
Action: Sets the diagonal elements of $\Sigma_{LD}$ to 0.¶
Potential usefulness: In updates to ldsc, will speed up estimation.¶
bash 2.1_mtag_no_overlap.bash¶
!python ../mtag/mtag.py \
--sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \
--out ../output/2_special_opts/2.1_mtag_no_overlap_EA \
--no_overlap \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.2 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --stream-stdout \ --no-overlap \ --sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \ --out ../output/2_special_opts/2.1_mtag_no_overlap_EA Beginning MTAG analysis... Read in Trait 1 summary statistics (8146840 SNPs) from ../data/EducAtt_ea2.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 8146840 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 8146840 SNPs remain. Removed 0 SNPs with duplicated rs numbers (8146840 SNPs remain). Removed 4434138 SNPs with N < 74074.074074 (3712702 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.85 Lambda GC = 1.627 Max chi^2 = 106.943 5297 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:06:09 2017 Total time elapsed: 38.55s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 3712702 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (1191004 SNPs) from ../data/EducAtt_ukb.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 1191004 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 12960 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 1178044 SNPs remain. Removed 0 SNPs with duplicated rs numbers (1178044 SNPs remain). Removed 0 SNPs with N < 35242.0 (1178044 SNPs remain). Median value of SIGNED_SUMSTATS was -0.000292861940766, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.205 Lambda GC = 1.192 Max chi^2 = 39.518 23 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:06:37 2017 Total time elapsed: 5.74s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 1178044 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 790847 SNPs remaining merging with previous traits. Dropped 1 SNPs due to inconsistent allele pairs from phenotype 2. 790846 SNPs remain. Flipped the signs of of 384828 SNPs to make them consistent with the effect allele orderings of the first trait. ... Merge of GWAS summary statistics complete. Number of SNPs: 790846 Using 790846 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 0.92 0. ] [ 0. 1.028]] Beginning estimation of Omega ... Using GMM estimator of Omega .. Checking for positive definiteness .. matrix is not positive definite, performing adjustment.. Completed in 0 iterations Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 2.501 371136 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 2.468 380236 Estimated Omega: [[ 6.492e-06 5.026e-06] [ 5.026e-06 3.970e-06]] Estimated Sigma: [[ 0.92 0. ] [ 0. 1.028]] MTAG results saved to file. MTAG complete. Time elapsed: 3.0m:1.71074914932s
!tail -26 ../output/2_special_opts/2.1_mtag_no_overLap_EA.log
2017/10/18/06:07:40 PM Checking for positive definiteness .. 2017/10/18/06:07:40 PM matrix is not positive definite, performing adjustment.. 2017/10/18/06:07:40 PM Completed in 0 iterations 2017/10/18/06:07:40 PM Completed estimation of Omega ... 2017/10/18/06:07:40 PM Beginning MTAG calculations... 2017/10/18/06:07:41 PM ... Completed MTAG calculations. 2017/10/18/06:07:41 PM Writing Phenotype 1 to file ... 2017/10/18/06:07:58 PM Writing Phenotype 2 to file ... 2017/10/18/06:08:15 PM Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 2.501 371136 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 2.468 380236 Estimated Omega: [[ 6.492e-06 5.026e-06] [ 5.026e-06 3.970e-06]] Estimated Sigma: [[ 0.92 0. ] [ 0. 1.028]] 2017/10/18/06:08:15 PM 2017/10/18/06:08:15 PM MTAG results saved to file. 2017/10/18/06:08:15 PM MTAG complete. Time elapsed: 3.0m:1.71074914932s
--perfect_gencov: Different measures of the same trait¶
Assumes: All summary statistics in MTAG are GWAS estimates for traits perfectly correlated with another.¶
Action: $\Omega$ is equal to outer product of the square root of the vector of genetic variances.¶
- This pins the genetic correlations to be 1.
Potential usefulness: MTAG on GWAS of different measures of the same trait.¶
Note: It is sometimes the case that different measures of the same trait are in fact not perfectly correlated (see Meghan Zacher's talk later today).¶
bash 2.2_perfect_gencov.bash¶
# 2.2_perfect_gencov.bash
!python ../mtag/mtag.py \
--sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \
--out ../output/2_special_opts/2.2_mtag_perf_gencov_EA \
--perfect_gencov \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.3 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --stream-stdout \ --perfect-gencov \ --sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \ --out ../output/2_special_opts/2.2_mtag_perf_gencov_EA Beginning MTAG analysis... Read in Trait 1 summary statistics (8146840 SNPs) from ../data/EducAtt_ea2.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 8146840 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 8146840 SNPs remain. Removed 0 SNPs with duplicated rs numbers (8146840 SNPs remain). Removed 4434138 SNPs with N < 74074.074074 (3712702 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.85 Lambda GC = 1.627 Max chi^2 = 106.943 5297 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:18:25 2017 Total time elapsed: 39.93s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 3712702 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (1191004 SNPs) from ../data/EducAtt_ukb.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 1191004 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 12960 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 1178044 SNPs remain. Removed 0 SNPs with duplicated rs numbers (1178044 SNPs remain). Removed 0 SNPs with N < 35242.0 (1178044 SNPs remain). Median value of SIGNED_SUMSTATS was -0.000292861940766, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.205 Lambda GC = 1.192 Max chi^2 = 39.518 23 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:18:52 2017 Total time elapsed: 5.76s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 1178044 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 790847 SNPs remaining merging with previous traits. Dropped 1 SNPs due to inconsistent allele pairs from phenotype 2. 790846 SNPs remain. Flipped the signs of of 384828 SNPs to make them consistent with the effect allele orderings of the first trait. ... Merge of GWAS summary statistics complete. Number of SNPs: 790846 Using 790846 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 0.92 0.385] [ 0.385 1.028]] Beginning estimation of Omega ... Using GMM estimator of Omega .. Checking for positive definiteness .. Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 2.022 252528 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 2.022 264572 Estimated Omega: [[ 6.492e-06 5.077e-06] [ 5.077e-06 3.970e-06]] Estimated Sigma: [[ 0.92 0.385] [ 0.385 1.028]] MTAG results saved to file. MTAG complete. Time elapsed: 3.0m:0.128737211227s
!tail -26 ../output/2_special_opts/2.2_mtag_perf_gencov_EA.log
2017/10/18/06:19:54 PM Beginning estimation of Omega ... 2017/10/18/06:19:54 PM Using GMM estimator of Omega .. 2017/10/18/06:19:54 PM Checking for positive definiteness .. 2017/10/18/06:19:54 PM Completed estimation of Omega ... 2017/10/18/06:19:54 PM Beginning MTAG calculations... 2017/10/18/06:19:56 PM ... Completed MTAG calculations. 2017/10/18/06:19:56 PM Writing Phenotype 1 to file ... 2017/10/18/06:20:12 PM Writing Phenotype 2 to file ... 2017/10/18/06:20:28 PM Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 2.022 252528 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 2.022 264572 Estimated Omega: [[ 6.492e-06 5.077e-06] [ 5.077e-06 3.970e-06]] Estimated Sigma: [[ 0.92 0.385] [ 0.385 1.028]] 2017/10/18/06:20:28 PM 2017/10/18/06:20:28 PM MTAG results saved to file. 2017/10/18/06:20:28 PM MTAG complete. Time elapsed: 3.0m:0.128737211227s
Compare:¶
| Analysis | $\Omega$ | Genetic correlation |
|---|---|---|
| Regular | [6.492e-06 4.815e-06] [4.815e-06 3.970e-06] |
[1.000 0.949] [0.949 1.000] |
| No overlap | [6.492e-06 5.026e-06] [5.026e-06 3.970e-06] |
[1.000 0.990] [0.990 1.000] |
| Perfect genetic correlation | [6.492e-06 5.077e-06] [5.077e-06 3.970e-06] |
[1.000 1.000] [1.000 1.000] |
--equal_h2: Performing meta-analysis with mtag¶
Action: Skips calculation Omega, uses simplified MTAG estimator¶
bash 2.3_mtag_equal_h2.bash¶
!python ../mtag/mtag.py \
--sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \
--out ../output/2_special_opts/2.3_mtag_perf_gencov_EA \
--perfect_gencov \
--equal_h2 \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.3 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --equal-h2 \ --stream-stdout \ --perfect-gencov \ --sumstats ../data/EducAtt_ea2.txt,../data/EducAtt_ukb.txt \ --out ../output/2_special_opts/2.3_mtag_perf_gencov_EA Beginning MTAG analysis... Read in Trait 1 summary statistics (8146840 SNPs) from ../data/EducAtt_ea2.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 8146840 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 8146840 SNPs remain. Removed 0 SNPs with duplicated rs numbers (8146840 SNPs remain). Removed 4434138 SNPs with N < 74074.074074 (3712702 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.85 Lambda GC = 1.627 Max chi^2 = 106.943 5297 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:39:18 2017 Total time elapsed: 35.26s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 3712702 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (1191004 SNPs) from ../data/EducAtt_ukb.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 1191004 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 12960 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 1178044 SNPs remain. Removed 0 SNPs with duplicated rs numbers (1178044 SNPs remain). Removed 0 SNPs with N < 35242.0 (1178044 SNPs remain). Median value of SIGNED_SUMSTATS was -0.000292861940766, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.205 Lambda GC = 1.192 Max chi^2 = 39.518 23 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:39:47 2017 Total time elapsed: 6.26s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 1178044 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 790847 SNPs remaining merging with previous traits. Dropped 1 SNPs due to inconsistent allele pairs from phenotype 2. 790846 SNPs remain. Flipped the signs of of 384828 SNPs to make them consistent with the effect allele orderings of the first trait. ... Merge of GWAS summary statistics complete. Number of SNPs: 790846 Using 790846 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 0.92 0.385] [ 0.385 1.028]] Beginning estimation of Omega ... --perfect_gencov and --equal_h2 option used Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 1.998 246761 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 1.998 258530 Omega hat not computed because --equal_h2 was used. Estimated Sigma: [[ 0.92 0.385] [ 0.385 1.028]] MTAG results saved to file. MTAG complete. Time elapsed: 3.0m:0.847772121429s
!tail -26 ../output/2_special_opts/2.3_mtag_perf_gencov_EA.log
2017/10/18/06:40:53 PM Checking for positive definiteness .. 2017/10/18/06:40:53 PM Sigma hat: [[ 0.92 0.385] [ 0.385 1.028]] 2017/10/18/06:40:53 PM Beginning estimation of Omega ... 2017/10/18/06:40:53 PM --perfect_gencov and --equal_h2 option used 2017/10/18/06:40:53 PM Completed estimation of Omega ... 2017/10/18/06:40:53 PM Beginning MTAG calculations... 2017/10/18/06:40:54 PM ... Completed MTAG calculations. 2017/10/18/06:40:54 PM Writing Phenotype 1 to file ... 2017/10/18/06:41:10 PM Writing Phenotype 2 to file ... 2017/10/18/06:41:27 PM Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 .../EducAtt_ea2.txt 250000 134825 790846 2.011 1.998 246761 2 .../EducAtt_ukb.txt 52863 52863 790846 1.204 1.998 258530 Omega hat not computed because --equal_h2 was used. Estimated Sigma: [[ 0.92 0.385] [ 0.385 1.028]] 2017/10/18/06:41:27 PM 2017/10/18/06:41:27 PM MTAG results saved to file. 2017/10/18/06:41:27 PM MTAG complete. Time elapsed: 3.0m:0.847772121429s
3. maxFDR calculations¶
--fdr: Calculates an approximate upper bound on the FDR based on the framework presented.
- Additional options for:
- Customized set of grid points to calculate FDR (
--grid_file) - Adjusting coarseness of grid (
--intervals) - Parallelization (
--cores) - Sample size approximation (
--n_approx) - Restrict grid points to that those that maintain the prior probability of a null SNP for each trait (in development).
- Customized set of grid points to calculate FDR (
- Default options calculate uses
--intervals 10and a single core.
3.1_max_FDR_basics.bash¶
# 3.1_max_FDR_basics.bash
!python ../mtag/mtag.py \
--sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \
--out ../output/3_maxFDR/3.1_mtag_maxFDR_basic_NS \
--stream_stdout \
--fdr \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.3 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --stream-stdout \ --sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \ --fdr \ --out ../output/3_maxFDR/3.1_mtag_maxFDR_basic_NS Beginning MTAG analysis... Read in Trait 1 summary statistics (185458 SNPs) from ../data/1_OA2016_hm3samp_NEUR.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 185458 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 185458 SNPs remain. Removed 0 SNPs with duplicated rs numbers (185458 SNPs remain). Removed 162680 SNPs with N < 74074.074074 (22778 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.467 Lambda GC = 1.366 Max chi^2 = 59.523 20 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:44:01 2017 Total time elapsed: 0.63s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 22778 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (167911 SNPs) from ../data/1_OA2016_hm3samp_SWB.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 167911 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 167911 SNPs remain. Removed 0 SNPs with duplicated rs numbers (167911 SNPs remain). Removed 102537 SNPs with N < 74074.074074 (65374 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.208 Lambda GC = 1.188 Max chi^2 = 29.353 1 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 18:44:03 2017 Total time elapsed: 0.68s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 65374 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 20321 SNPs remaining merging with previous traits. ... Merge of GWAS summary statistics complete. Number of SNPs: 20321 Using 20321 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 1.043 -0.129] [-0.129 0.966]] Beginning estimation of Omega ... Using GMM estimator of Omega .. Checking for positive definiteness .. Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 ...hm3samp_NEUR.txt 111111 111111 20321 1.407 1.488 133309 2 ..._hm3samp_SWB.txt 111111 111111 20321 1.269 1.408 168490 Estimated Omega: [[ 3.817e-06 -2.199e-06] [ -2.199e-06 2.336e-06]] Estimated Sigma: [[ 1.043 -0.129] [-0.129 0.966]] MTAG results saved to file. Beginning maxFDR calculations. Depending on the number of grid points specified, this might take some time... T=2 Number of gridpoints to search: 65 Performing grid search using 1 cores. Grid search: 10.0 percent finished for . Time: 0.004 min Grid search: 20.0 percent finished for . Time: 0.005 min Grid search: 30.0 percent finished for . Time: 0.007 min Grid search: 40.0 percent finished for . Time: 0.008 min Grid search: 50.0 percent finished for . Time: 0.009 min Grid search: 60.0 percent finished for . Time: 0.010 min Grid search: 70.0 percent finished for . Time: 0.011 min Grid search: 80.0 percent finished for . Time: 0.011 min Grid search: 90.0 percent finished for . Time: 0.012 min Grid search: 100.0 percent finished for . Time: 0.013 min Saved calculations of fdr over grid points in ../output/3_maxFDR/3.1_mtag_maxFDR_basic_NS_fdr_mat.txt <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> grid point indices for max FDR for each trait: [12 3] Maximum FDR Max FDR of Trait 1: 0.00109655427181 at probs = [ 0. 0.3 0. 0.7] Max FDR of Trait 2: 0.00436959625046 at probs = [ 0. 0. 0.3 0.7] <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Completed FDR calculations. MTAG complete. Time elapsed: 25.1162800789s
!tail -26 ../output/3_maxFDR/3.1_mtag_maxFDR_basic_NS.log
2017/10/18/06:44:25 PM 2017/10/18/06:44:25 PM MTAG results saved to file. 2017/10/18/06:44:25 PM Beginning maxFDR calculations. Depending on the number of grid points specified, this might take some time... 2017/10/18/06:44:25 PM T=2 2017/10/18/06:44:25 PM Number of gridpoints to search: 65 2017/10/18/06:44:25 PM Performing grid search using 1 cores. 2017/10/18/06:44:25 PM Grid search: 10.0 percent finished for . Time: 0.004 min 2017/10/18/06:44:25 PM Grid search: 20.0 percent finished for . Time: 0.005 min 2017/10/18/06:44:25 PM Grid search: 30.0 percent finished for . Time: 0.007 min 2017/10/18/06:44:25 PM Grid search: 40.0 percent finished for . Time: 0.008 min 2017/10/18/06:44:25 PM Grid search: 50.0 percent finished for . Time: 0.009 min 2017/10/18/06:44:25 PM Grid search: 60.0 percent finished for . Time: 0.010 min 2017/10/18/06:44:25 PM Grid search: 70.0 percent finished for . Time: 0.011 min 2017/10/18/06:44:25 PM Grid search: 80.0 percent finished for . Time: 0.011 min 2017/10/18/06:44:25 PM Grid search: 90.0 percent finished for . Time: 0.012 min 2017/10/18/06:44:25 PM Grid search: 100.0 percent finished for . Time: 0.013 min 2017/10/18/06:44:25 PM Saved calculations of fdr over grid points in ../output/3_maxFDR/3.1_mtag_maxFDR_basic_NS_fdr_mat.txt 2017/10/18/06:44:25 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> 2017/10/18/06:44:25 PM grid point indices for max FDR for each trait: [12 3] 2017/10/18/06:44:25 PM Maximum FDR 2017/10/18/06:44:25 PM Max FDR of Trait 1: 0.00109655427181 at probs = [ 0. 0.3 0. 0.7] 2017/10/18/06:44:25 PM Max FDR of Trait 2: 0.00436959625046 at probs = [ 0. 0. 0.3 0.7] 2017/10/18/06:44:25 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> 2017/10/18/06:44:25 PM Completed FDR calculations. 2017/10/18/06:44:25 PM MTAG complete. Time elapsed: 25.1162800789s
The number of grid points rises dramatically in the number of traits and inverse size of intervals!¶
In this case, we recommend that you use --n_approx¶
--n_approx Speed up FDR calculation by replacing the sample size
of a SNP for each trait by the mean across SNPs (for
each trait). Recommended.bash 3.2_maxFDR_approx.bash¶
!python ../mtag/mtag.py \
--sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \
--out ../output/3_maxFDR/3.2_mtag_maxFDR_approx_NS \
--stream_stdout \
--fdr \
--intervals 100 \
--n_approx \
--stream_stdout
<><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> <> MTAG: Multitrait Analysis of GWAS <> Version: 1.0.3 <> (C) 2017 Omeed Maghzian, Raymond Walters, and Patrick Turley <> Harvard University Department of Economics / Broad Institute of MIT and Harvard <> GNU General Public License v3 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> <> Note: It is recommended to run your own QC on the input before using this program. <> Software-related correspondence: maghzian@nber.org <> All other correspondence: paturley@broadinstitute.org <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Calling ./mtag.py \ --intervals 100 \ --stream-stdout \ --n-approx \ --sumstats ../data/1_OA2016_hm3samp_NEUR.txt,../data/1_OA2016_hm3samp_SWB.txt \ --fdr \ --out ../output/3_maxFDR/3.2_mtag_maxFDR_approx_NS Beginning MTAG analysis... Read in Trait 1 summary statistics (185458 SNPs) from ../data/1_OA2016_hm3samp_NEUR.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 1 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 185458 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 185458 SNPs remain. Removed 0 SNPs with duplicated rs numbers (185458 SNPs remain). Removed 162680 SNPs with N < 74074.074074 (22778 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.467 Lambda GC = 1.366 Max chi^2 = 59.523 20 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 19:17:06 2017 Total time elapsed: 0.77s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 1 complete. SNPs remaining: 22778 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Read in Trait 2 summary statistics (167911 SNPs) from ../data/1_OA2016_hm3samp_SWB.txt ... <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging Trait 2 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><>< <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Interpreting column names as follows: snpid: Variant ID (e.g., rs number) n: Sample size a1: Allele 1, interpreted as ref allele for signed sumstat. pval: p-Value a2: Allele 2, interpreted as non-ref allele for signed sumstat. freq: Allele frequency z: Directional summary statistic as specified by --signed-sumstats. Reading sumstats from provided DataFrame into memory 10000000 SNPs at a time. . done Read 167911 SNPs from --sumstats file. Removed 0 SNPs with missing values. Removed 0 SNPs with INFO <= None. Removed 0 SNPs with MAF <= 0.01. Removed 0 SNPs with out-of-bounds p-values. Removed 0 variants that were not SNPs. Note: strand ambiguous SNPs were not dropped. 167911 SNPs remain. Removed 0 SNPs with duplicated rs numbers (167911 SNPs remain). Removed 102537 SNPs with N < 74074.074074 (65374 SNPs remain). Median value of SIGNED_SUMSTATS was 0.0, which seems sensible. Dropping snps with null values Metadata: Mean chi^2 = 1.208 Lambda GC = 1.188 Max chi^2 = 29.353 1 Genome-wide significant SNPs (some may have been removed by filtering). Conversion finished at Wed Oct 18 19:17:08 2017 Total time elapsed: 0.98s <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Munging of Trait 2 complete. SNPs remaining: 65374 <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Trait 2 summary statistics: 20321 SNPs remaining merging with previous traits. ... Merge of GWAS summary statistics complete. Number of SNPs: 20321 Using 20321 SNPs to estimate Omega (0 SNPs excluded due to strand ambiguity) Estimating sigma.. Checking for positive definiteness .. Sigma hat: [[ 1.043 -0.129] [-0.129 0.966]] Beginning estimation of Omega ... Using GMM estimator of Omega .. Checking for positive definiteness .. Completed estimation of Omega ... Beginning MTAG calculations... ... Completed MTAG calculations. Writing Phenotype 1 to file ... Writing Phenotype 2 to file ... Summary of MTAG results: ------------------------ Trait N (max) N (mean) # SNPs used GWAS mean chi^2 MTAG mean chi^2 GWAS equiv. (max) N 1 ...hm3samp_NEUR.txt 111111 111111 20321 1.407 1.488 133309 2 ..._hm3samp_SWB.txt 111111 111111 20321 1.269 1.408 168490 Estimated Omega: [[ 3.817e-06 -2.199e-06] [ -2.199e-06 2.336e-06]] Estimated Sigma: [[ 1.043 -0.129] [-0.129 0.966]] MTAG results saved to file. Beginning maxFDR calculations. Depending on the number of grid points specified, this might take some time... T=2 Number of gridpoints to search: 33580 Performing grid search using 1 cores. Grid search: 10.0 percent finished for . Time: 0.214 min Grid search: 20.0 percent finished for . Time: 0.442 min Grid search: 30.0 percent finished for . Time: 0.674 min Grid search: 40.0 percent finished for . Time: 0.905 min Grid search: 50.0 percent finished for . Time: 1.134 min Grid search: 60.0 percent finished for . Time: 1.357 min Grid search: 70.0 percent finished for . Time: 1.581 min Grid search: 80.0 percent finished for . Time: 1.794 min Grid search: 90.0 percent finished for . Time: 2.048 min Grid search: 100.0 percent finished for . Time: 2.276 min Saved calculations of fdr over grid points in ../output/3_maxFDR/3.2_mtag_maxFDR_approx_NS_fdr_mat.txt <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> grid point indices for max FDR for each trait: [ 829 33576] Maximum FDR Max FDR of Trait 1: 0.00110246986219 at probs = [ 0. 0.28 0. 0.72] Max FDR of Trait 2: 0.0271483484174 at probs = [ 0.97 0. 0.01 0.02] <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> Completed FDR calculations. MTAG complete. Time elapsed: 3.0m:6.61157798767s
!tail -26 ../output/3_maxFDR/3.2_mtag_maxFDR_approx_NS.log
2017/10/18/07:17:30 PM 2017/10/18/07:17:30 PM MTAG results saved to file. 2017/10/18/07:17:30 PM Beginning maxFDR calculations. Depending on the number of grid points specified, this might take some time... 2017/10/18/07:17:30 PM T=2 2017/10/18/07:17:54 PM Number of gridpoints to search: 33580 2017/10/18/07:17:54 PM Performing grid search using 1 cores. 2017/10/18/07:18:07 PM Grid search: 10.0 percent finished for . Time: 0.214 min 2017/10/18/07:18:21 PM Grid search: 20.0 percent finished for . Time: 0.442 min 2017/10/18/07:18:35 PM Grid search: 30.0 percent finished for . Time: 0.674 min 2017/10/18/07:18:49 PM Grid search: 40.0 percent finished for . Time: 0.905 min 2017/10/18/07:19:02 PM Grid search: 50.0 percent finished for . Time: 1.134 min 2017/10/18/07:19:16 PM Grid search: 60.0 percent finished for . Time: 1.357 min 2017/10/18/07:19:29 PM Grid search: 70.0 percent finished for . Time: 1.581 min 2017/10/18/07:19:42 PM Grid search: 80.0 percent finished for . Time: 1.794 min 2017/10/18/07:19:57 PM Grid search: 90.0 percent finished for . Time: 2.048 min 2017/10/18/07:20:11 PM Grid search: 100.0 percent finished for . Time: 2.276 min 2017/10/18/07:20:12 PM Saved calculations of fdr over grid points in ../output/3_maxFDR/3.2_mtag_maxFDR_approx_NS_fdr_mat.txt 2017/10/18/07:20:12 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> 2017/10/18/07:20:12 PM grid point indices for max FDR for each trait: [ 829 33576] 2017/10/18/07:20:12 PM Maximum FDR 2017/10/18/07:20:12 PM Max FDR of Trait 1: 0.00110246986219 at probs = [ 0. 0.28 0. 0.72] 2017/10/18/07:20:12 PM Max FDR of Trait 2: 0.0271483484174 at probs = [ 0.97 0. 0.01 0.02] 2017/10/18/07:20:12 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><> 2017/10/18/07:20:12 PM Completed FDR calculations. 2017/10/18/07:20:12 PM MTAG complete. Time elapsed: 3.0m:6.61157798767s
Output for when using the option is relevant¶
2017/09/28/05:21:04 PM Calculating FDR by setting number of intervals to 10, and True use of approximation.
2017/09/28/05:21:04 PM T=2
2017/09/28/05:21:04 PM Number of gridpoints to search: 92
2017/09/28/05:21:04 PM Performing grid search using 20 cores.
2017/09/28/05:21:05 PM Grid search: 10.0 percent finished for . Time: 0.006 min
2017/09/28/05:21:05 PM Grid search: 20.0 percent finished for . Time: 0.011 min
2017/09/28/05:21:06 PM Grid search: 30.0 percent finished for . Time: 0.016 min
2017/09/28/05:21:06 PM Grid search: 40.0 percent finished for . Time: 0.022 min
2017/09/28/05:21:06 PM Grid search: 50.0 percent finished for . Time: 0.027 min
2017/09/28/05:21:06 PM Grid search: 60.0 percent finished for . Time: 0.032 min
2017/09/28/05:21:07 PM Grid search: 70.0 percent finished for . Time: 0.037 min
2017/09/28/05:21:07 PM Grid search: 80.0 percent finished for . Time: 0.043 min
2017/09/28/05:21:07 PM Grid search: 90.0 percent finished for . Time: 0.048 min
2017/09/28/05:21:08 PM Grid search: 100.0 percent finished for . Time: 0.054 min
2017/09/28/05:21:08 PM Saved calculations of fdr over grid points in /var/genetics/ukb/huili/mtag/output/main/fdr_calculation/interval_10_True_meanapprox_fdr_mat.txt
2017/09/28/05:21:08 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
2017/09/28/05:21:08 PM grid point indices for max FDR for each trait: [90 3]
2017/09/28/05:21:08 PM Maximum FDR
2017/09/28/05:21:08 PM Max FDR of Trait 1: 0.104708870765 at probs = [ 0.8 0.1 0. 0.1]
2017/09/28/05:21:08 PM Max FDR of Trait 2: 0.000149109160478 at probs = [ 0. 0. 0.3 0.7]
2017/09/28/05:21:08 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
2017/09/28/05:21:08 PM Completed FDR calculations.
2017/09/28/05:21:08 PM Calculating FDR by setting number of intervals to 10, and False use of approximation.
2017/09/28/05:21:08 PM T=2
2017/09/28/05:21:08 PM Number of gridpoints to search: 92
2017/09/28/05:21:08 PM Performing grid search using 20 cores.
2017/09/28/05:22:30 PM Grid search: 10.0 percent finished for . Time: 1.371 min
2017/09/28/05:23:52 PM Grid search: 20.0 percent finished for . Time: 2.743 min
2017/09/28/05:25:15 PM Grid search: 30.0 percent finished for . Time: 4.112 min
2017/09/28/05:26:37 PM Grid search: 40.0 percent finished for . Time: 5.480 min
2017/09/28/05:27:59 PM Grid search: 50.0 percent finished for . Time: 6.858 min
2017/09/28/05:29:22 PM Grid search: 60.0 percent finished for . Time: 8.230 min
2017/09/28/05:30:44 PM Grid search: 70.0 percent finished for . Time: 9.599 min
2017/09/28/05:32:07 PM Grid search: 80.0 percent finished for . Time: 10.978 min
2017/09/28/05:33:29 PM Grid search: 90.0 percent finished for . Time: 12.350 min
2017/09/28/05:34:52 PM Grid search: 100.0 percent finished for . Time: 13.728 min
2017/09/28/05:34:52 PM Saved calculations of fdr over grid points in /var/genetics/ukb/huili/mtag/output/main/fdr_calculation/interval_10_False_meanapprox_fdr_mat.txt
2017/09/28/05:34:52 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
2017/09/28/05:34:52 PM grid point indices for max FDR for each trait: [90 3]
2017/09/28/05:34:52 PM Maximum FDR
2017/09/28/05:34:52 PM Max FDR of Trait 1: 0.104764850463 at probs = [ 0.8 0.1 0. 0.1]
2017/09/28/05:34:52 PM Max FDR of Trait 2: 0.000149016665847 at probs = [ 0. 0. 0.3 0.7]
2017/09/28/05:34:52 PM <><><<>><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><><>
2017/09/28/05:34:52 PM Completed FDR calculations.Workflow for running maxFDR¶
Run MTAG in the background, work with results while maxFDR is still running.¶
Questions?¶
Ways to get help:
- Check out the tutorials available on the Github wiki
- Refer back to the original MTAG paper (especially the Supplementary Note)
- For troubleshooting, post a message on the Google Group
- If you find any bugs/errors, open up an issue on Github
- Finally, email: maghzian@nber.org
Conclusion¶
MTAG allows analysis of GWAS of multiple correlated traits using summary statistics
Identifies more loci and improves polygenic prediction for depressive symptoms, neuroticism, and subjective well-being
Code is publicly available at: https://github.com/omeed-maghzian/mtag